Today’s Medicinal Chemist – a checklist for Candidate selection
By Steve Djuric
Many (many) years ago as a fledgling medicinal chemist I was instructed to find compounds with good potency, bioavailability and in vivo pharmacology. Having done that, I was told that I was well on the way to candidate selection. Well, clearly, things have changed in the intervening years and the current stellar crop of medicinal chemists need to identify compounds with the following properties amongst others:
- Excellent potency for target
- General adherence to Lipinski type properties apart from bRo5 (1), PPI, TF and PROTACS etc classes of compound
- Well-designed BEI, LipE, LigE, LLE, LELP, LipMetE (2) etc characteristics
- Acceptable bioavailability
- Avoidance of metabolic hotspots leading to activation and idiosyncratic toxicity
- Often tissue/organ selectivity
- Acceptable clearance of free drug
- Manageable CYP inhibition
- No CYP induction liability
- Limited efflux potential especially for CNS targeted compounds
- Good safety including hERG, dog CV and 7-day rodent safety
- IVIV correlations for PK. Determination of clearance mechanisms. PBPK modeling for dose prediction
- Demonstration of target engagement in cells and in vivo (Pfizer pillars of success (3))
- Determination of levels of free drug needed for efficacy vs toxicity Determination of a Therapeutic Index in a few animal species
- Excellent selectivity – let’s take a quick look at this. We do CEREP or Eurofins screens, kinome profiling on hundreds of kinases (often against isolated enzymes only instead of in a relevant cellular context! (4)) and convince ourselves that compounds are selective (often after generating a few IC50s for off targets identified in the selectivity panel to convince ourselves that there is a significant window). Then we run dog CV studies or 7-day rodent toxicology studies and find a problem. We conclude that the candidate was not selective and must be hitting an off target. Some then turn to CETSA, PISA, Eurofins InCELL assays (5) etc and then discuss what that data means! Recent advances include transcriptomics/connectivity maps (6). Otherwise, the solution is to just make more compounds.
Interestingly, several startup companies have argued with me that you really don’t need all this information in a development candidate package. Are they right?! All on a case-by-case basis, perhaps?
As this is a DMPK blog let’s look at the PK demands more closely. I included several of them in the list above.
Permeability of PROTACs
We want to evaluate oral absorption, CNS penetration, cell permeability etc. The Permeability/Efflux – PAMPA, Caco2 and MDCK/MDR1 – workhorse assays generally do a fair job. However, in the current Discovery environment where we work on many projects involving bRo5 compounds and protein degraders/PROTACs (now including tri-functional PROTACs! (7)), we struggle to predict/assess permeability of compounds. Despite the excellent bRo5 guidelines provided by DeGoey, Cox et al. at AbbVie cited above, we have little guidance on how PROTAC type compounds get into cells. Given MWs of frequently over 1000, one might argue against passive permeability yet many workers feel that absorption is passive but kinetically slow. Others, including Kell at Manchester (8) argue that most/all molecules are actively transported. The question remains as to what is being done to help chemists design more permeable PROTACs. What rules are being developed? What mechanistic studies are being done? The answer cannot be simply to ask chemists to keep making more molecules until they find something that works. Current PROTACs demonstrate a wide range of cell permeabilities and oral bioavailability. In the context of BRo5 and PROTACs, a recent paper has suggested that while cell based in vitro tools of permeability may predict medium and high permeability, they have limited utility for low, and the best bet is rodent PO/PK studies (a very nice data analysis looking at pre-clinical approaches compared to human oral absorption) (9).
Dealing with CYP metabolism
We started with being asked to avoid DDI potential of compounds. So, we look for metabolism by CYPs (or CYP450 inhibition). It’s 3A4, then 3A4 and 2D6 that one needs to be careful of then 3A4, 2D6, 1A2 and so on. Then, the DMPK experts tell use to avoid hitting just one of these enzymes as they have candidate development ability concerns and would we please produce compounds with acceptable clearance of free drug that are cleared by multiple CYPs. A huge part of this concern is the increased use of combination therapies as we tackle more challenging diseases. It’s one thing to develop a standalone molecule that may have DDI with co-meds, it’s very different if you need to combine two meds right from the get go and their delivery is sometimes simultaneous.
OK – so how, as a medicinal chemist, do you design all that in? Or is it trial and error again?
What about aldehyde oxidase?
Moreover, more recently, we find that a number of heterocycles are targeted not so much by CYPs but by aldehyde oxidase. Aldehyde oxidase (AO) catalyzes oxidations of azaheterocycles and aldehydes, amide hydrolysis, and diverse reductions. Challenges here arise from complex and poorly understood AO biology, which hinders an effective solution in stopping or decreasing AO metabolism. Part of the reason that DMPK colleagues discourage it, is there is often little organizational tolerance for failure due to PK at the SAD/MAD phase and we just aren’t good at predicting human PK accurately when it comes to complex AO metabolism.
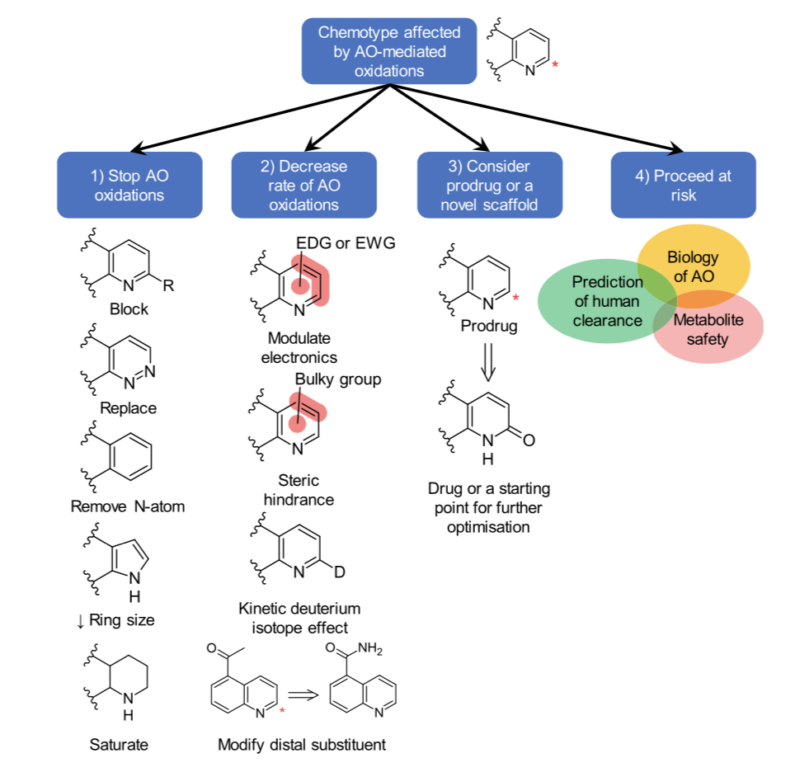
Figure reprinted with permission from Manevski et al. Metabolism by Aldehyde Oxidase: Drug Design and Complementary Approaches to Challenges in Drug Discovery. J. Med. Chem. 2019, 62, 24, 10955–10994. https://doi.org/10.1021/acs.jmedchem.9b00875. Copyright 2019 American Chemical Society
An excellent recent perspective has focused on rational drug design approaches to modulate AO‐mediated metabolism in drug discovery (10). The article also reflects on the computational design of compounds that avoid AO.
Impact of AI
So, where am I going with all this? If one looks at the medicinal chemist check list for the properties of optimal candidate compounds it is huge. Are all these criteria necessary, desirable? Is it possible to rationally design compounds that fulfill the criteria outlined, or are we still in an era where despite huge advances in drug design (compound properties, in silico computational approaches including ML/AI etc etc), we are still reduced to taking as many shots on goal as possible?
Recently, AI/ML approaches have been claimed to generate optimized chemical matter more efficiently, particularly for tractable targets (InSilico Medicine, ChemPass and Exscientia (11) to name but a few). De-novo generative design platforms can be configured to optimize many properties concurrently using ML based automated MPO (Multiparameter Optimization) algorithms, and indeed some companies claim to be getting compounds in the clinic much faster and more efficiently (significantly less compounds per candidate) using this approach. The one caveat to AI/ML is that it is unlikely to work on less tractable targets like PPIs, but for kinases and other enzymes and even GPCRs these platforms are becoming mainstream. Having said this, the AI/ML community suggest that to optimize many properties at once is routinely possible as long as the data is robust, and the models are reasonably accurate. Looking at the multi-CYP selectivity problem and off target identification challenges mentioned above, amongst others – is this really so at the moment?
Certainly, in terms of selectivity/off targets, it seems that we have a long way to go and one wonders if this is actually doable? Perhaps Connectivity maps may ultimately help?
Where do we go from here?
So, ending on an optimistic note we can anticipate that:
- De-novo generative design platforms can be configured to optimize many properties concurrently using DL based automated MPO algorithms. As they become more sophisticated/accurate (improved training sets), they should provide significant help in the area of MPO of candidate molecules. Including prediction of PK properties (12) This is particularly true in the realm of traditional/known protein family targets.
- Coupled with High Throughput Experimentation techniques (13), integrated synthesis- purification-bioassay platforms (14) should be able to get real time bioassay data on compounds that have been prioritized for synthesis by AI platforms.
- Connectivity maps using different cell lines, CETSA, PISA etc should help us identify off targets of our compounds. Transcriptional profiling may also help predict drug efficacy (15).
- From a pragmatic and bottom-line perspective, if a is compound safe and potent and can reach the target effectively, a lot of the other issues go away. PK experts often push for little to no DDI because at the large doses often required, it’s nearly impossible to avoid DDI, with the same being true with idiosyncratic toxicity – it rears its head at high dose. For sure, this is sometimes easier said than done with bRo5 compounds, but with the exquisite potency of PROTACs it isn’t uncommon to see doses < 100 mgs.
What do folks think? Let’s discuss…
Stevan W Djuric, PhD
President, Discovery Chemistry and Technology Consulting LLC
Acknowledgements:
Thanks to Drs Kelly Desino and Phil Cox (AbbVie) and Dr Jim Summers (St. Andrews Consulting LLC) for valuable and insightful comments (some incorporated into text above) on content.
References
- David A. DeGoey, Hui-Ju Chen, Philip B. Cox, and Michael D. Wendt. Med. Chem. 2018, 61, 2636−2651; https://doi.org/10.1021/acs.jmedchem.7b00717
- Antonia F. Stepan, Gregory W. Kauffman, Christopher E. Keefer, Patrick R. Verhoest, and Martin Edwards. J. Med. Chem. 2013, 56, 6985−6990 and references therein; https://doi.org/10.1021/jm4008642
- Paul Morgan, Piet H. Van Der Graaf, John Arrowsmith, Doug E. Feltner, Kira S. Drummond, Craig D. Wegner and Steve D.A. Street. Drug Discovery Today, 2012, 17, Issues 9–10, 419-424; https://doi.org/10.1016/j.drudis.2011.12.020
- https://www.kinativ.com/
- Stina Lundgren. ACS Med. Chem. Lett. 2019, 10, 690−693; https://doi.org/10.1021/acsmedchemlett.9b00112. Massimiliano Gaetani, Pierre Sabatier, Amir A. Saei, Christian M. Beusch, Zhe Yang, Susanna L. Lundström, and Roman A. Zubarev. Proteome Res. 2019, 18, 4027−4037; https://doi.org/10.1021/acs.jproteome.9b00500. https://www.discoverx.com/products-applications/target-engagement-assays.
- Aravind Subramanian, Rajiv Narayan, Steven M. Corsello, …, David E. Root, Bang Wong, Todd R. Golub. Cell, 2017, 171, 1437-1452.E17; https://doi.org/10.1016/j.cell.2017.10.049.
- Satomi Imaide, Kristin M. Riching , Nikolai Makukhin, Vesna Vetma, Claire Whitworth, Scott J. Hughes, Nicole Trainor, Sarah D. Mahan, Nancy Murphy, Angus D. Cowan ,Kwok-Ho Chan , Conner Craigon, Andrea Testa, Chiara Maniaci , Marjeta Urh ,Danette L. Daniels and Alessio Ciull Nat. Chem. Biol. 17, 1157–1167 (2021); https://doi.org/10.1038/s41589-021-00878-4.
- Paul D. Dobson, Karin Lanthaler, Stephen G Oliver, and Douglas B Kell. Curr. Top Med. Chem., 2009, 9(2), 163-81; doi: 2174/156802609787521616
- Edward Price, J. Cory Kalvass, David DeGoey, Balakrishna Hosmane, Stella Doktor,and Kelly Desino. J. Med. Chem., 2021, 64, 13, 9389–9403; https://doi.org/10.1021/acs.jmedchem.1c00669.
- Nenad Manevski, Lloyd King, William R. Pitt, Fabien Lecomte, and Francesca Toselli, J. Med. Chem. 2019, 62, 10955−10994; https://doi.org/10.1021/acs.jmedchem.9b00875. David J. St. Jean, Jr., and Christopher Fotsch. J. Med. Chem., 2012, 55, 6002−6020; https://doi.org/10.1021/jm300343m.
- Some interesting thoughts on the ExScientia communications from Derek Lowe’s In the Pipeline blog https://www.science.org/content/blog-post/ai-generated-clinical-candidates-so-far
- Filip Miljkovic,́ Anton Martinsson, Olga Obrezanova, Beth Williamson, Martin Johnson, Andy Sykes, Andreas Bender, and Nigel Greene, Mol. Pharmaceutics 2021; https://doi.org/10.1021/acs.molpharmaceut.1c00718
- See, for example, Noah P. Tu, Amanda W. Dombrowski,Gashaw M. Goshu, Anil Vasudevan, Stevan W. Djuric , Ying Wang. Angew. Chem. Int. Ed. 2019, 58, 7987; https://doi.org/10.1002/anie.201900536 and references therein
- Aleksandra Baranczak, Noah P Tu, Jasmina Marjanovic, Philip A Searle, Anil Vasudevan, and Stevan W Djuric, ACS Med. Chem. Lett. 2017, 8, 4, 461-465; https://doi.org/10.1021/acsmedchemlett.7b00054.
- Jie Zhu, Jingxiang Wang, Xin Wang, Mingjing Gao, Bingbing Guo, Miaomiao Gao, Jiarui Liu, Yanqiu Yu, Liang Wang, Weikaixin Kong, Yongpan An, Zero Liu, Xinpei Sun, Zhuo Huang , Hong Zhou, Ning Zhang, Ruimao Zheng and Zhengwei Xie. Nat. Biotechnol. 39, 1444–1452 (2021); https://doi.org/10.1038/s41587-021-00946-z