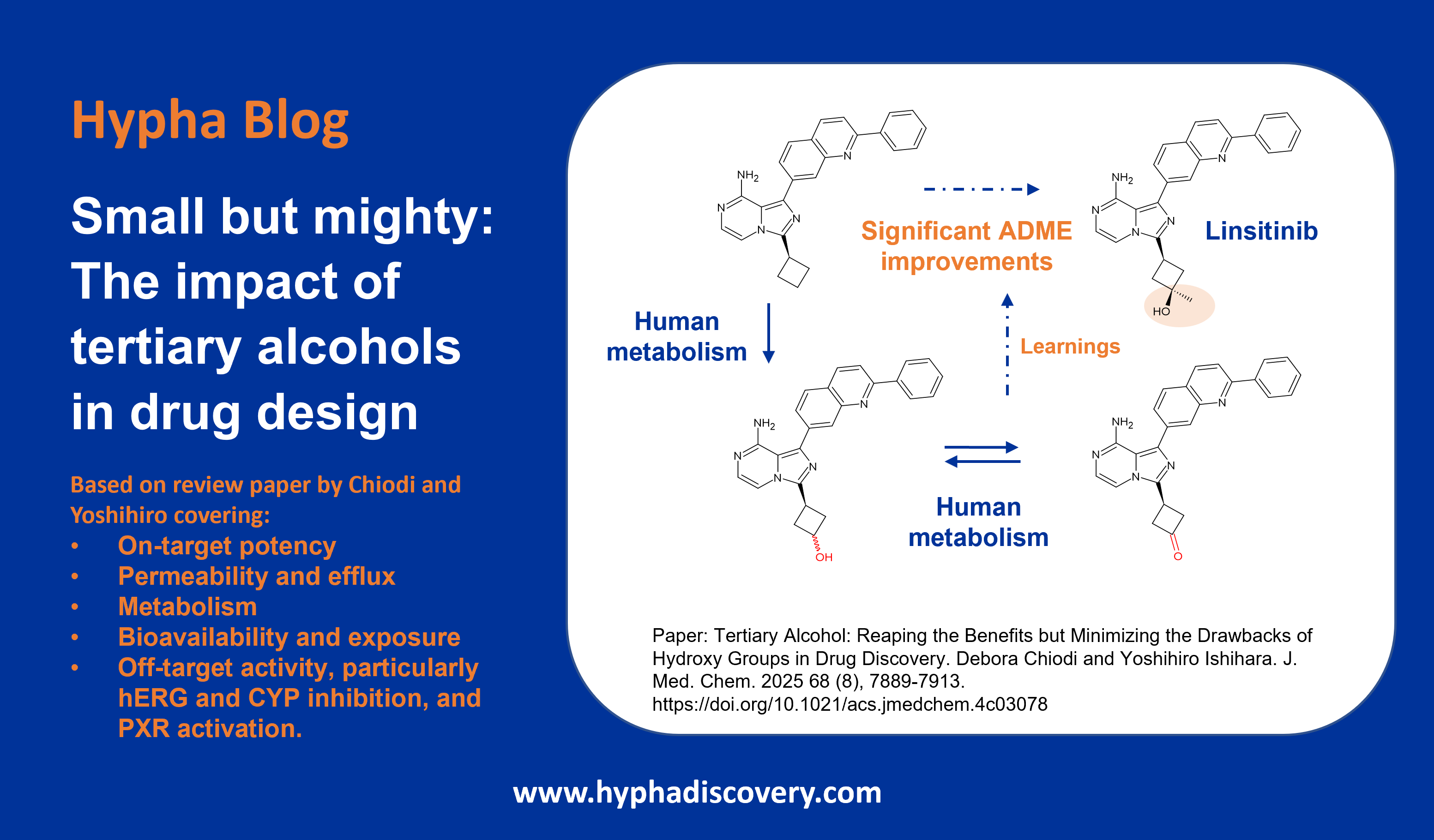
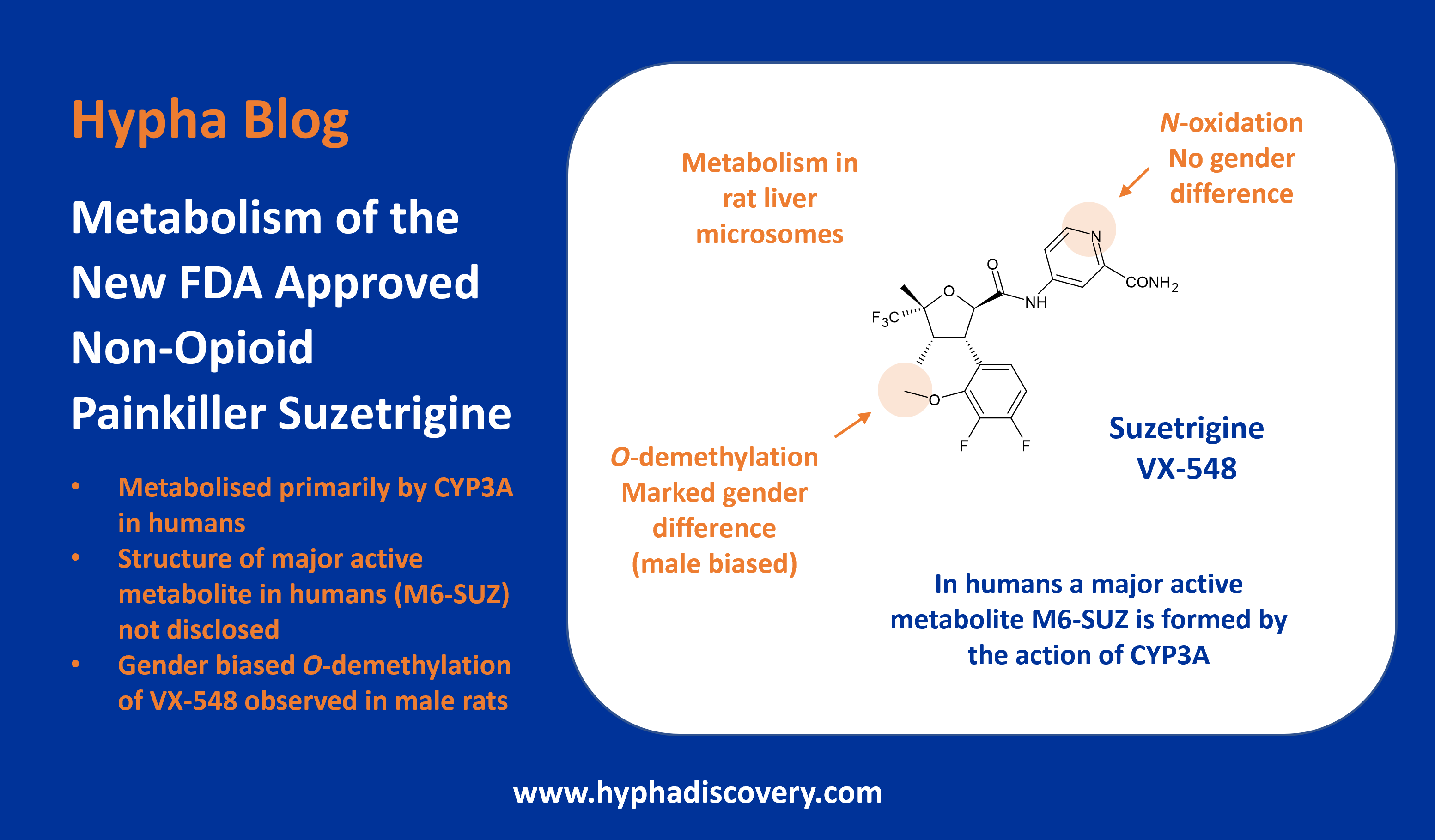
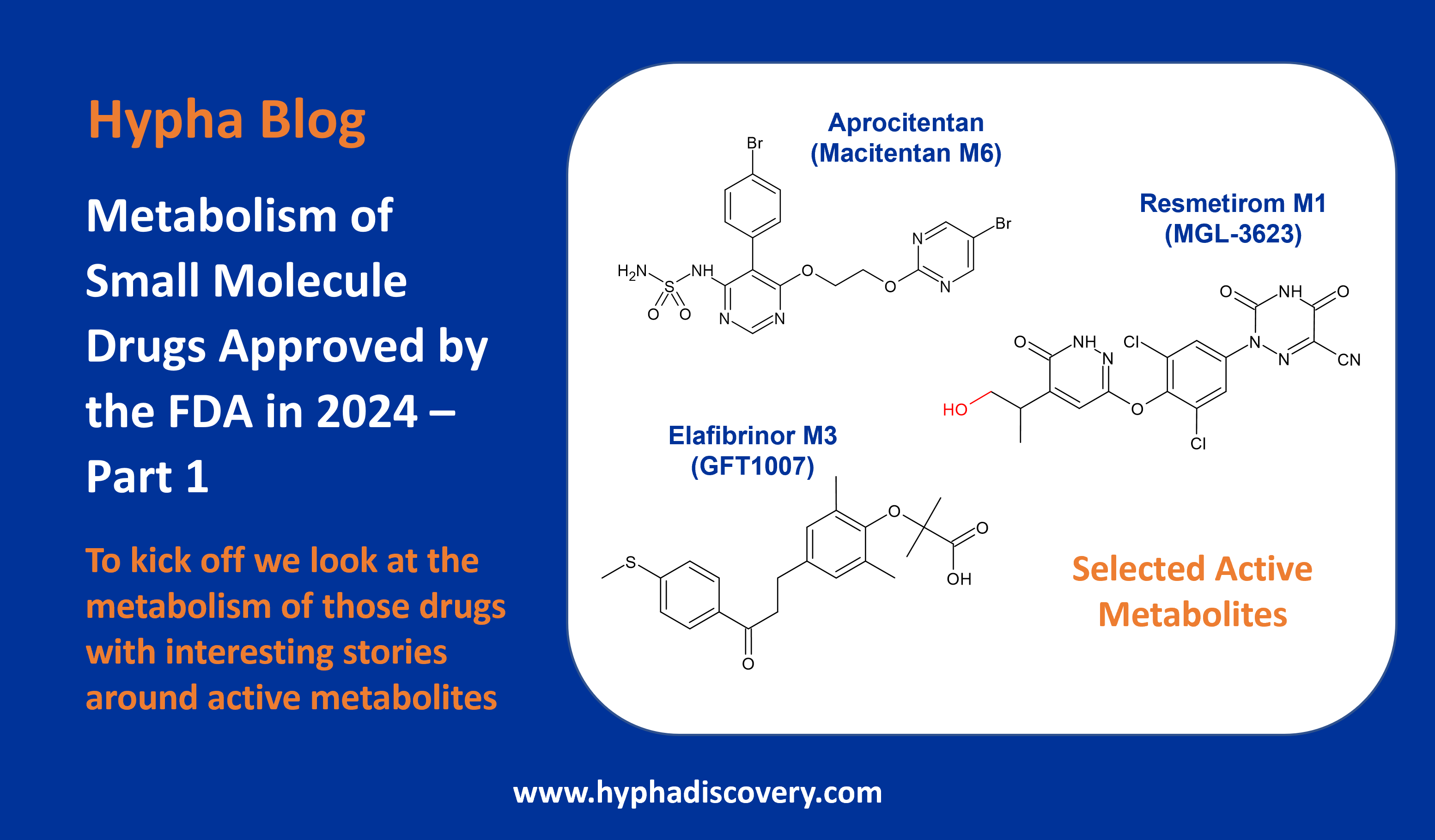
LipMetE – A useful tool for relating lipophilicity to metabolic stability in microsomes?
By Samuel Coe
LipMetE is a potentially useful and underutilised efficiency metric that relates logD to microsomal clearance. In this post we explore its origins and application to analyse metabolism trends in drug design.
High human liver microsome clearance is associated with sub optimal pharmacokinetics as well as high doses to achieve effective concentrations in vivo. These factors combine to increase the risk for adverse safety events in clinical trials.1 Lipophilic compounds (clogP>3) are often predisposed to have high microsomal clearance due to the lipophilic character of P450 binding sites.2
LipMetE was first reported by scientists at Pfizer while investigating cycloalkyl ether metabolism, describing it as the Yin to the Yang of LipE.3
While LipE is given by the equation:
LipE = -log10(Ki or IC50) – logD
and is often displayed graphically to show the efficiency of the potency of a compound relative to its lipophilicity (Figure 1).
LipMetE can also be displayed graphically but is defined by the equation:
LipMetE = logD – log10(CLint,u)
Where:
CLint, u = CLint,app/fu,mic
Whereas data for intrinsic clearance (CLint) in human liver microsomes is routinely collected in drug discovery projects, the fraction unbound in microsomes (fu,mic), is not a parameter consistently collected due to its labour and time intensive nature. Fortunately, in silco models can be an alternative and for those without the benefit of in-house models, Hallifax and Houston amongst others have published empirical equations for calculating fraction unbound in microsomes.4
The Pfizer article has garnered attention with nearly 50 citations since publication. The original report provides some interesting insights into how best to use the metric, which we would like to highlight in this blog. We will then attempt to apply it to some open source data from the literature.
Interpreting a LipMetE plot
Using data from the supporting information we can construct then examine the features of a LipMetE plot. Similar to a LipE plot, the compounds with the same LipMetE value will cluster around the same LipMetE line. For example, the compounds in the orange box are clustered around the LipMetE = 1 line. Moving up and down this line likely indicates lipophilicity as the major influence on the metabolism (Figure 2).

Figure 2 – LipMetE Plot with data from supporting information
For those compounds of equivalent LogD moving between LipMetE lines is indicative of a structural change, resulting in reduced affinity for the enzyme or a blocking of a metabolic soft spot (Figure 3). In this example, the gem-dimethyl oxetane products produced a positive gain in LipMetE predicting a blocking of a metabolic soft spot that was identified in an earlier publication.5

Figure 3 – LipMetE Plot highlighting outliers
Perhaps that most valuable result to the drug discovery community as a whole was the deep dive the authors took into the Pfizer data archives.

Figure 4 – THP matched pair transformation
To establish if LipMetE could be applied generally and not just to this small collection of g-secretase inhibitors, they mined the collection for various molecular matched pairs of cyclic ethers.
They found 113 matched pairs that when changing from a 3-THP to a 4-THP (Figure 4), produced an average net gain in LipMetE of 0.13 arising from almost equivalent logDs with reduced HLM. This is a valuable piece of knowledge for the medicinal chemist and more of these analyses would be very useful.
Applying LipMetE to the literature
In an attempt to apply LipMetE to a “hot topic” we identified an open source paper containing HLM Clint data for a collection of phenyl bioisosteres. One of the perceived benefits of bioisosteres is improved physiochemical properties relative to the phenyl ring6 and LipMetE should allow us to test this hypothesis by interrogating how changes in logD influence the metabolism.
The data comes from the Open Source Malaria group, who recently published a series of novel anti-malarials.7 Unfortunately, many of the phenyl replacements had reduced potency against the 3D7 strain of P. falciparum and were not characterised further. Taking the available data, we have calculated the LipMetE values for the compounds with reported HLM Clint (using the Hallifax and Houston method for calculating fu,mic), and the outcome is insightful (Figure 5).

Figure 5 – LipMetE plot
The Figure shows that the cubane, phenyl and carboranes analogues have similar LipMetE values, clustering around 2. The bicyclopentane (BCP) derivative meanwhile made a clear improvement in LipMetE with a value of 2.6. Applying the LipMetE hypothesis we could conclude that this improvement is attributed to the structural change because the logD is similar to the phenyl analogue.
In the article, the authors describe in detail a biofunctionalization pathway that leads to the higher HLM clearance of cubane. Investigations identified an unstable, hydroxylated intermediate, capable of degradation. While this is an excellent study, for others applying phenyl bioisosteres at an early stage in drug development programs it may simply be enough in future to use LipMetE analysis. Using LipMetE would identify the lack of improvement cubane offers in stability, for the lipophilicity gain and allows the program to move onto testing alternative isosteres.
As the article contains many other non-classical phenyl bioisosteres in the same chemical series. It would have been interesting if the HLM Clint data for these had been available. This could have revealed other interesting observations in the LipMetE analysis, possibly providing knowledge for the wider community.
Do you use LipMetE?
Despite LipMetE being a potentially useful tool to analyse metabolism trends, we note that it isn’t often reported in the literature and are curious to know how often it is applied. Do you have a large data set that could be mined for trends such as the 3-THP to a 4-THP transformation? Have you used LipMetE in programs but not published the results? We would love to hear your thoughts and comments.
References
1 Physiochemical drug properties associated with in vivo toxicological outcomes. Hughes, J. D.; Blagg, J.; Price, D. A.; Bailey, S.; DeCrescenzo, G. A.; Devraj, R. V.; Ellsworth, E.; Fobian, Y. M.; Gibbs, M. E.; Gilles, R. W.; Greene, N.; Huang, E.; Krieger-Burke, T.; Loesel, J.; Wager, T.; Whiteley, L.; Zhang, Y. Bioorg. Med. Chem. Lett., 2008, 18, 4872−4875.
2 Baseline lipophilicity relationships in human cytochromes P450 associated with drug metabolism. Lewis, D. F.; Dickins, M. Drug Metab. Rev., 2003, 35, 1−18.
3 Evaluating the Differences in Cycloalkyl Ether Metabolism Using the Design Parameter “Lipophilic Metabolism Efficiency” (LipMetE) and a Matched Molecular Pairs Analysis. Stepan, A. F.; Kauffman, G. W.; Keefer, C. E.; Verhoest, P. R.; Edwards, M. J. Med. Chem., 2013, 56, 6985−6990.
4 Binding of drugs to hepatic microsomes: comment and assessment of current prediction methodology with recommendation for improvement. Hallifax, D.; Houston, J. B. Drug Metab. Dispos., 2006, 34, 724−726.
5 Metabolism-Directed Design of Oxetane-Containing Arylsulfonamide Derivatives as γ-Secretase Inhibitors. Stepan, A. F.; Karki, K.; McDonald, W. S.; Dorff, P. H.; Dutra, J. K.; DiRico, K. J.; Won, A.; Subramanyam, C.; Efremov, I. V.; O’Donnell, C. J.; Nolan, C. E.; Becker, S. L.; Pustilnik, L. R.; Sneed, B.; Sun, H.; Lu, Y.; Robshaw, A. E.; Riddell, D.; O’Sullivan, T. J.; Sibley, E.; Capetta, S.; Atchison, K.; Hallgren, A. J.; Miller, E.; Wood, A.; Obach, R. S. J. Med. Chem., 2011, 54, 7772−7783.
6 Bioisosteres of the Phenyl Ring: Recent Strategic Applications in Lead Optimization and Drug Design. Subbaiah, M. A. M.; Meanwell, N. A. J. Med. Chem., 2021, 64, 14046−14128 (and references contained within).
7 Nonclassical phenyl bioisosteres as effective replacements in a series of novel open-source antimalarials. Tse, E. G.; Houston, S. D.; Williams, C. M.; Savage, G. P.; Rendina, L. M.; Hallyburton, I.; Anderson, M.; Sharma, R.; Walker, G. S.; Obach, R. S.; Todd, M. H. J. Med. Chem., 2020, 63, 11585−11601.