Metabolism of covalent drugs: Stories from a resurgent field
By Samuel Coe
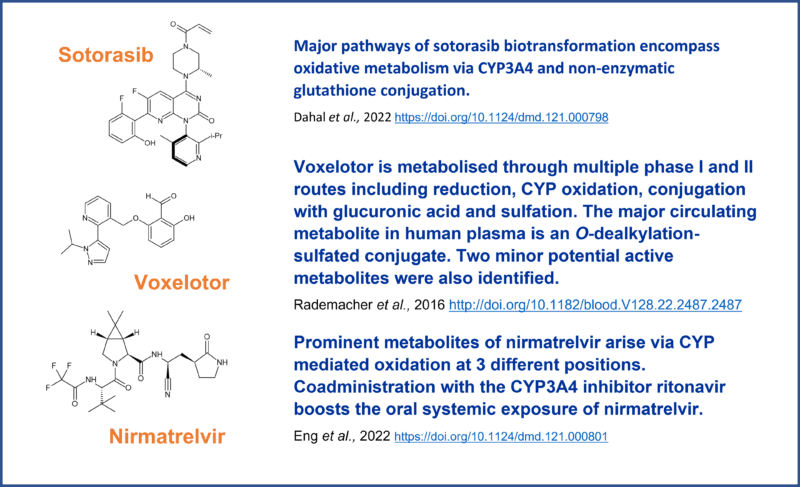
With the successful launch of Amgen’s targeted KRAS-blocking lung cancer drug sotorasib, Pfizer’s nirmatrelvir antiviral and GBT’s (now Pfizer) sickle cell anemia drug voxelotor (Figure 1), it is clear that over the past decade, covalent drugs have seen a revival in popularity. This is largely attributed to advances in rational drug design that has seen a new class of covalent compound emerge. The so called “targeted covalent inhibitors” combine structure-based design for warheads with high selectivity but low promiscuous reactivity.
Covalent inhibitors are typically small molecules with a mildly reactive functional group that forms a covalent bond with targeted proteins.1a They bind on a spectrum from reversible to effectively irreversible and are ideally suited to niches where complete inhibition/destruction of the target is required (cancer/anti-infectives), or where the natural ligand is present in high concentrations (kinases). However, such properties require careful balancing of parameters such as dose to limit toxicity.1a,b
Throughout this article we highlight some interesting metabolism quirks of covalent inhibitors. Although metabolism by cytochrome P450s is still a major pathway for covalent compounds there are many non-CYP pathways such as direct conjugation with glutathione that can have a large impact.2
The early days
Besides aspirin, penicillins are the earliest widely used covalent inhibitors. Their mechanism of action involves ring opening of a strained beta-lactam ring by DD-transpeptidase, preventing bacterial cell wall cross-linking. Covalent inhibitors are generally believed to be less susceptible to resistance but in the case of penicillin, specific beta-lactamases have led to early penicillins being less effective.3
In the early 20th century, drugs were sometimes brought to market before the mechanism of action was fully known but have since been reclassified as having a covalent mode of action.4 One of most widely prescribed drugs, the proton pump inhibitor omeprazole, works through formation of a disulfide linkage to cysteine residues in the H+/K+ ATPase pump in gastric secretory cells. Under the acidic conditions of the stomach, the free sulfenic acid can form that, after loss of water, results in the more labile sulfenamide cyclised product (Figure 2). This N-S bond can react irreversibly with a free cysteine, inhibiting the ability of the proton pump H+/K+ ATPase to secrete gastric acid. Recently it has been shown that omeprazole binds to a wide range of proteins, which is presumably the reason for the associated unwanted effects that can arise from long term use of this proton pump inhibitor.5
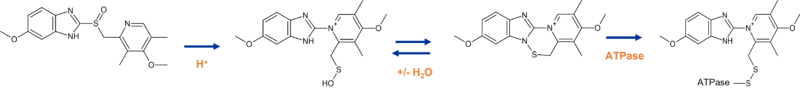
Figure 2 Omeprazole mechanism of action
Greater selectivity
As the field of rational and structure-based drug design grew, it became apparent that rather than using covalent inhibitors as sacrificial agents (as is the case with penicillins and omeprazole), there could be an opportunity to first develop a highly selective molecule, then take advantage of a nearby cysteine residue to covalently bind to the pocket. This approach was particularly attractive to kinase targets where selectivity can be achieved by design, but due to the high concentration of the ATP natural ligand, it can be difficult to convert the selectivity to efficacy in vivo.
Because of their inherent reactivity, it is recommended that covalent inhibitors are screened at an early stage for metabolic activation via cytochrome P450 enzymes (CYPs).6 This can be achieved with in vitro trapping experiments such as liver microsome incubations in the presence of glutathione (GSH), with and without NADPH, and N-α-acetyl-l-lysine amine model assays.7 There are also non-CYP mediated bioactivation pathways that can be identified with chemical proteomics techniques and an example is given later on. These offer a way to improve the knowledge around off targets effects in the future.
Although the majority of targeted covalent inhibitors to reach the market have been generally free of overt metabolic activation leading to toxicity, ibrutinib is one exception.8
Bioactivation of ibrutinib
The major metabolite of ibrutinib is formed through epoxidation of the acrylamide warhead by CYP3A4, resulting in a 1,2 dihydrodiol (Figure 3).
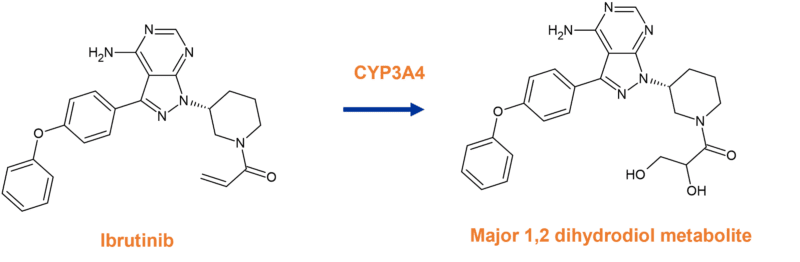
Figure 3 Major metabolism pathway of ibrutinib to a 1,2 dihydrodiol metabolite
While this may be the source of the observed toxicities by alkylation of off-target proteins, an additional extra hepatic pathway has been shown to be the cause of kidney toxicity.8
After GSH conjugation of ibrutinib via the action of glutathione S-transferases, the resulting adduct is excreted in two ways; either via the bile into faeces, or in urine via the kidney. In doing so, cysteine conjugates are generated that result in toxic effects, sometimes leading to kidney failure.
In the proposed pathway deconvoluted by Rood et. al., formation of cysteine-metabolites by renal dipeptidases could accelerate the bioactivation pathway through cysteine-S-conjugate ß-lyase, an enzyme expressed in renal tubules often involved in bioactivation and nephrotoxicity. This enzyme is able to cleave the cystine residue, resulting in a thiol group in the pharmacophore. The thiol of ibrutinib-SH is then able to form the unwanted protein conjugates, or is inactivated by formation of other conjugates e.g. glucuronides (Figure 4).8
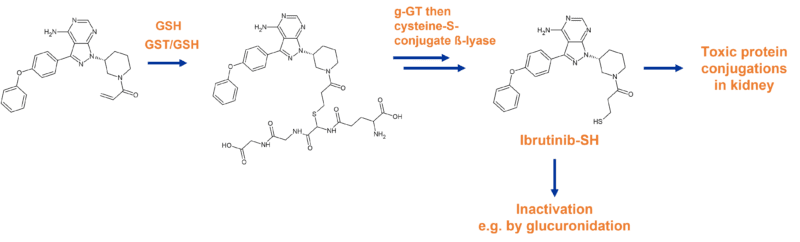
Figure 4 Abridged pathway of ibrutinib biotransformation resulting in toxic conjugates
Mammalian cysteine-S-conjugate ß-lyases are mainly amino-acid metabolizing enzymes that have ß-lyase activity and cause the unwanted side reaction. While most studies on the enzymes have focused on bioactivation of halogenated alkenes, studies such as the one above highlights the potential of ß-lyase activity in the metabolism of other covalent motifs.8
BIA 10-2474 and the impact on aldehyde dehydrogenase-2
As mentioned earlier, proteomics is an excellent tool for identifying potentially harmful off-target interactions. The activity-based protein profiling involves modifying the compound of interest with a reporting tag (Figure 5). Tags can come in a number of forms e.g. biotin, photoaffinity etc. After incubation and pull down, it is possible to identify proteins that have been modified by the covalent inhibitors, providing valuable information.9
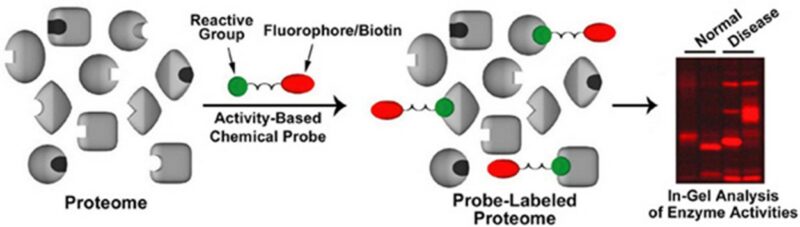
Figure 5 Schematic of chemical probe technology (source https://www.scripps.edu/cravatt/research.html)
This technique was used to great effect whilst investigating the neurotoxicity seen in a Phase I clinical trial of BIA 10–2474, a fatty acid amide hydroxylase inhibitor. In the 2016 trial, one subject died and four were hospitalized with neurological symptoms. Until this point, fatty acid amide hydroxylase inhibitors had been well tolerated in the clinic and it was a suspected that an off-target activity was to blame.10
The team first synthesised the major circulating metabolites of BIA-10-2474 (BIA-10-2445, BIA-10-2583, and the more minor compound 4, illustrated in Figure 6) before modifying them with a “clickable” alkyne tag (compounds 5-8 in Figure 6). After confirming that this modification had little impact on the on-target activity, activity-based protein profiling experiments identified aldehyde dehydrogenase-2 (ALDH-2) as the inhibited enzyme. Aldehyde dehydrogenases are thought to protect the brain from reactive oxygen species and reactive aldehydes, but as they are cysteine-dependent, are susceptible to covalent modifications. Site-directed mutagenesis further confirmed the reactivity of the desmethyl metabolite BIA-10-2583.10
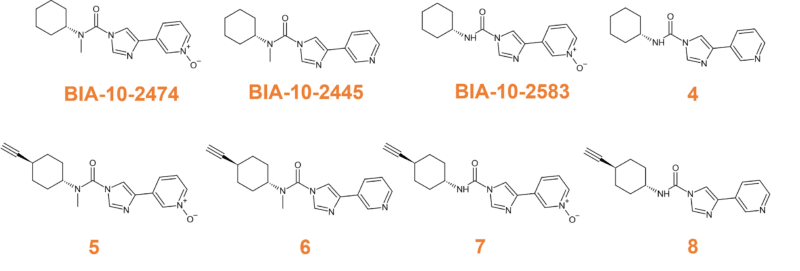
Figure 6
Additional proteomic experiments resolved the mechanism of inhibition. Briefly, initial N-demethylation produces a metabolite that once bound to ALDH-2, undergoes elimination of the imidazole forming a highly reactive isocyanate. The Cys319 residue subsequently reacts with the isocyanate and the modified enzyme is unable to carry out its normal function of protecting from neural damage (Figure 7).
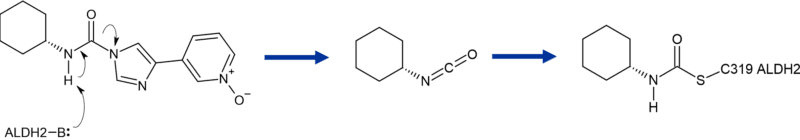
Figure 7 Cartoon of formation of reactive isocyanate from major metabolite BIA 10-2583 during biotransformation of BIA 10-2474
Metabolism of sotorasib
Coming full circle to sotorasib, there are indications that the mechanism of metabolism may develop into a story requiring extensive deconvolution.
Animal studies with 14C labelled compound identified 30 metabolites in rats and 11 in dogs. In both species’ glutathione conjugation was the primary metabolism pathway (M12) with other pathways being minor contributors to the overall metabolism picture11 (Figure 8 shows the definitive metabolites that have been chemically synthesised as standards).
Rats have also been shown to produce an additional sulphur containing metabolite that is a putative product of the cysteine S-conjugate β-lyase pathway.12 In humans however, the EMC submission document states hepatoxicity displaying as elevated levels of alanine aminotransferase and aspartate aminotransferase, with the main metabolic pathways being CYP3A4 oxidation and non-enzymatic conjugation with glutathione. It is also a mild inducer of CYP3A4.13
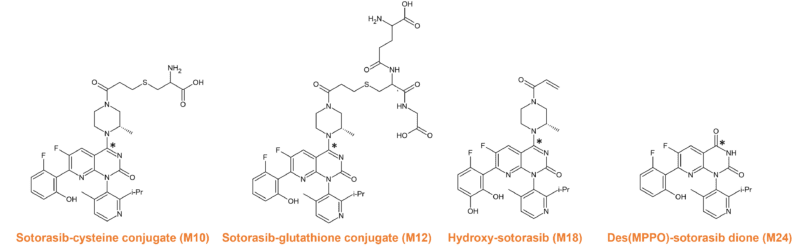
Figure 8 Synthesised metabolites of sotorasib. 14C indicated by *
A deeper understanding
Covalent inhibitors provide fascinating stories of drug discovery and their mechanisms of action are now more readily understood through the use of chemical biology techniques. This deeper understanding allows potential safety concerns to be addressed before the compounds are too far into the development pipeline and we imagine covalent inhibitors will continue to be an important modality for developing new medicines.
References
1a. Boike, L., Henning, N.J. & Nomura, D.K. Advances in covalent drug discovery. Nat Rev Drug Discov (2022). https://doi.org/10.1038/s41573-022-00542-z
1b. Singh, J. 2022 J. Med. Chem. 2022, 65, 8, 5886–5901. https://doi.org/10.1021/acs.jmedchem.1c02134
2. Park, B., Boobis, A., Clarke, S. et al., 2011. Managing the challenge of chemically reactive metabolites in drug development. Nat Rev Drug Discov10, 292–306. https://doi.org/10.1038/nrd3408
3. Péczka, N, Orgován,Z., Ábrányi-Balogh, P. & Keserű, G.M. (2022). Electrophilic warheads in covalent drug discovery: an overview, Expert Opinion on Drug Discovery, 17:4, 413-422. https://doi.org/10.1080/17460441.2022.2034783
4. Robertson, J., 2005. Mechanistic Basis of Enzyme-Targeted Drugs. Biochemistry 2005, 44, 15, 5561–5571. https://doi.org/10.1021/bi050247e
5. Cartee N.M.P., Wang M.M. (2020). Binding of omeprazole to protein targets identified by monoclonal antibodies. PLoS ONE 15(9): e0239464. https://doi.org/10.1371/journal.pone.0239464
6. Baillie, T.A. (2021). Approaches to mitigate the risk of serious adverse reactions in covalent drug design, Expert Opinion on Drug Discovery, 16:3, 275-287. https://doi.org/10.1080/17460441.2021.1832079
7. Dahal, U.P., Gilbert, A.M., Obach, R.S. et al., 2016. Intrinsic reactivity profile of electrophilic moieties to guide covalent drug design: N-α-acetyl-l-lysine as an amine nucleophile. Med. Chem. Commun., 2016,7, 864-872. DOI https://doi.org/10.1039/C6MD00017G
8. Rood, J.J.M., Jamalpoor, A., van Hoppe, S. et al., 2021. Extrahepatic metabolism of ibrutinib. Invest New Drugs 39, 1–14. https://doi.org/10.1007/s10637-020-00970-x
9. Shan, W, Yu, T., Min, W., et al., 2018. Advanced Activity-Based Protein Profiling Application Strategies for Drug Development. Frontiers in Pharmacology. https://www.frontiersin.org/articles/10.3389/fphar.2018.00353
10. Huang, Z., Ogasawara, D., Seneviratne, U.I., et al., 2019. Global Portrait of Protein Targets of Metabolites of the Neurotoxic Compound BIA 10–2474. ACS Chem. Biol. 2019, 14, 2, 192–197. https://doi.org/10.1021/acschembio.8b01097
11. Dahal, U.P., Rock, B.M., Rodgers, J., et al., 2022. ADME of sotorasib in rats and dogs. Drug Metabolism and Disposition February 13, 2022, DMD-AR-2021-000798; DOI: https://doi.org/10.1124/dmd.121.000798
12. Werner, J.A. Rhian Davies, R., Jan Wahlstrom, J., et al., 2021. Mercapturate pathway metabolites of sotorasib, a covalent inhibitor of KRASG12C, are associated with renal toxicity in the Sprague Dawley rat. Toxicology and Applied Pharmacology, 423, 115578. https://doi.org/10.1016/j.taap.2021.115578
13. EMC Lumykras (sotorasib). https://www.medicines.org.uk/emc/product/12871#PHARMACODYNAMIC_PROPS
References cited in Figure 1
ADME of Sotorasib in Rats and Dogs. Upendra P. Dahal, Brooke M. Rock, John Rodgers, Xiaomeng Shen, Zhe Wang and Jan L. Wahlstrom. Drug Metabolism and Disposition May 1, 2022, 50 (5) 600-612; DOI: https://doi.org/10.1124/dmd.121.000798
Absorption, Metabolism and Excretion of GBT440, a Novel Hemoglobin S (HbS) Polymerization Inhibitor for the Treatment of Sickle Cell Disease (SCD), in Healthy Male Subjects. Peter Rademacher, Athiwat Hutchaleelaha, Carla Washington, Josh Lehrer, Eleanor Ramos. Blood 2016; 128 (22): 2487. doi: https://doi.org/10.1182/blood.V128.22.2487.2487
Pharmacokinetics and Metabolism of Nirmatrelvir. Heather Eng, Alyssa L. Dantonio, Eugene P. Kadar, R. Scott Obach, Li Di, Jian Lin, Nandini C. Patel, Britton Boras, Gregory S. Walker, Jonathan J. Novak, Emi Kimoto, Ravi Shankar P. Singh and Amit S. Kalgutkar. Drug Metabolism and Disposition May 1, 2022, 50 (5) 576-590; DOI: https://doi.org/10.1124/dmd.121.000801